Research in biomedical text mining is starting to produce technology which can make information in biomedical literature more accessible for bio-scientists. The main goal of this group is developing effective extraction algorithm or tools to find out the hidden or potent bio-evidence from mass biomedical articles (i.e. PubMed). In the past, our developed algorithms and tools are addressing to biological entity interaction extraction and pathway network construction. Now, we are interested in documents ranking and summarization aimed at chemical-related articles. In the future, we also look forward to process other type of data, not only the biological documents.
Developed software
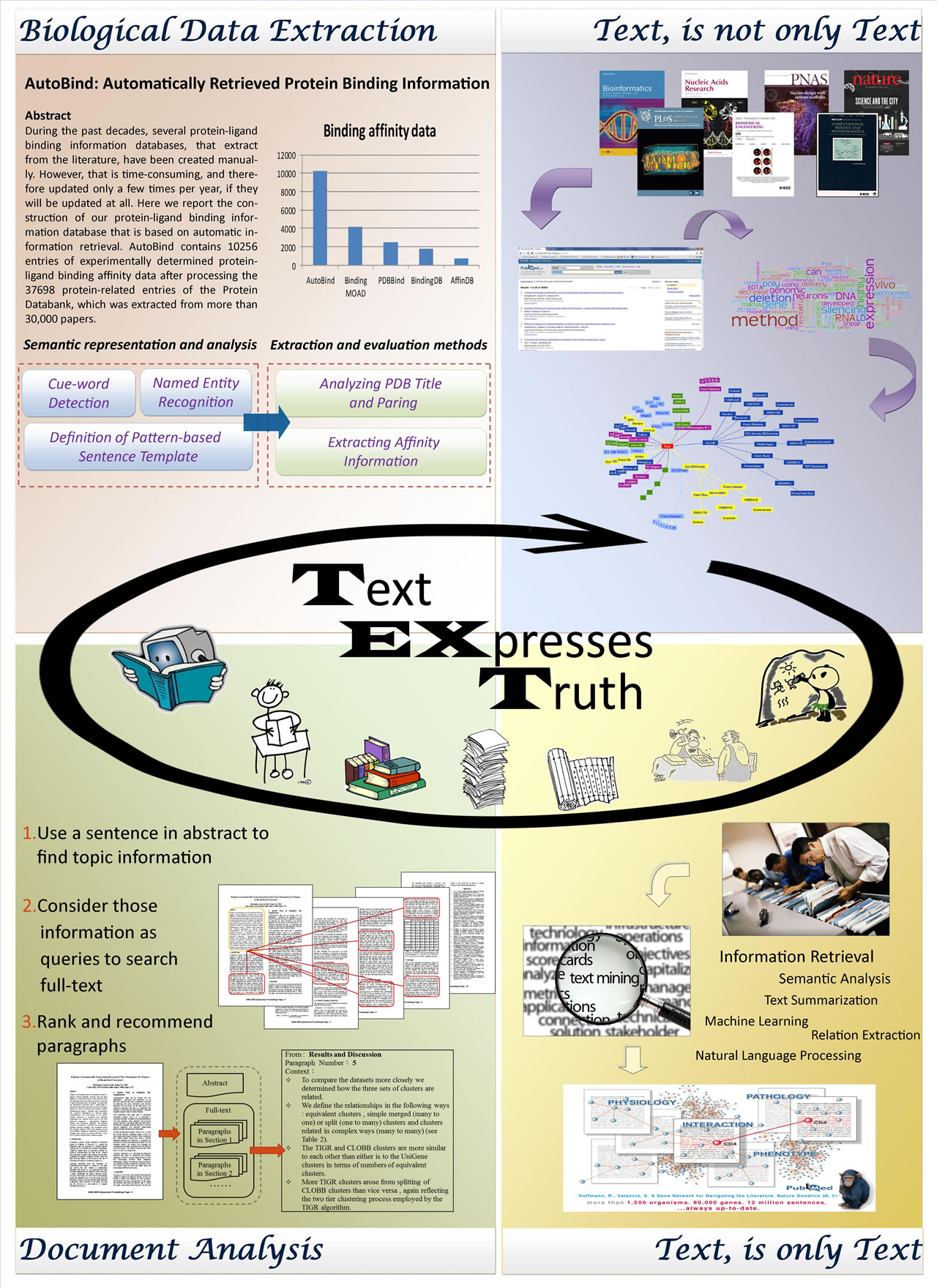
- AutoBind: Automatic extraction of protein-ligand binding affinity data from biological literature
http://autobind.csie.ncku.edu.tw - iExplorePathway: an intuitive web-based network exploration system for extending existing pathways using literature-based evidences
http://iexplorepathway.csie.ncku.edu.tw - Dr.TW: a biomedical term weighting method for document recommendation
http://140.116.247.184/~biocreative/ - C2HI: a Complete CHemical Information decision system
http://140.116.247.181/C2HI/ - CoTri: extracting chemical-disease relations with co-reference resolution and common trigger words
Download & Usage
Publication
- Darby Tien-Hao Chang, Chao-Hsuan Ke, Jung-Hsin Lin, and Jung-Hsien Chiang, AutoBind: Automatic extraction of protein-ligand binding affinity data from biological literature, Bioinformatics, Vol. 28 : 2162-2168, 2012.
DOI: http://dx.doi.org/10.1093/bioinformatics/BTS367
- Jung-Hsien Chiang, Heng-Hui Liu and Yi-Ting Huang, Condensing biomedical journal texts through paragraph ranking, Bioinformatics vol. 27 no. 8, 2011
DOI: http://dx.doi.org/10.1093/bioinformatics/BTR080
- Chao-Hsuan Ke, Tsung-Lu Michael Lee, Jung-Hsien Chiang (2012), C2HI: a Complete CHemical Information decision system, BioCreative 2012 Workshop, Washington, DC USA, Apr. 4-5, in press
- Jiun-Huang Ju, Yu-De Chen, Jung-Hsien Chiang, DrTW: A Biomedical Term Weighting Method for Document Recommendation, BioCreative 2012 Workshop, Washington, DC USA, Apr. 4-5, in press
- Jung-Hsien Chiang, Chao-Hsuan Ke, You-Cheng Lin, Ju-Ming Wang (2011), Extraction of signaling network from literature used for tracking novel route in pathway, The 5th Asian Young Researchers Conference on Computational and Omics Biology (AYRCOB 2011), Daejeon, Korea, Aug. 10-12, 76.
Recent Research Topic
